Hepatitis B virus
| Hepatitis B virus | |
|---|---|

| |
| Transmission electron microscopy micrograph showing Hepatitis B virus virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Pararnavirae |
| Phylum: | Artverviricota |
| Class: | Revtraviricetes |
| Order: | Blubervirales |
| Family: | Hepadnaviridae |
| Genus: | Orthohepadnavirus |
| Species: | Hepatitis B virus
|
Hepatitis B virus (HBV) is a partially double-stranded DNA virus,[1] a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses.[2][3] This virus causes the disease hepatitis B.[4]
Classification
[edit]Hepatitis B virus is classified in the genus Orthohepadnavirus, which contains 11 other species.[3] The genus is classified as part of the Hepadnaviridae family, which contains four other genera, Avihepadnavirus, Herpetohepadnavirus, Metahepadnavirus and Parahepadnavirus.[3] This family of viruses is the only member of the viral order Blubervirales.[3] Viruses similar to hepatitis B have been found in all apes (orangutans, gibbons, bonobos, gorillas and chimpanzees), in Old World monkeys,[5] and in New World woolly monkeys (the woolly monkey hepatitis B virus), suggesting an ancient origin for this virus in primates.
The virus is divided into four major serotypes (adr, adw, ayr, ayw) based on antigenic epitopes present on its envelope proteins. These serotypes are based on a common determinant (a) and two mutually exclusive determinant pairs (d/y and w/r). The viral strains have also been divided into ten genotypes (A–J) and forty subgenotypes according to overall nucleotide sequence variation of the genome.[6] The genotypes have a distinct geographical distribution and are used in tracing the evolution and transmission of the virus. Differences between genotypes affect the disease severity, course and likelihood of complications, and response to treatment.[7][8] The serotypes and genotypes do not necessarily correspond.
Genotype D has 10 subgenotypes.[9][6]
Unclassified species
[edit]A number of as yet unclassified hepatitis B-like species have been isolated from bats.[10]
Morphology
[edit]Structure
[edit]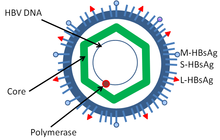
Hepatitis B virus is a member of the Hepadnavirus family.[11] The virus particle, called Dane particle[12] (virion), consists of an outer lipid envelope and an icosahedral nucleocapsid core composed of protein. The nucleocapsid encloses the viral DNA and a DNA polymerase that has reverse transcriptase activity similar to retroviruses.[13] The outer envelope contains embedded proteins which are involved in viral binding of, and entry into, susceptible cells. The virus is one of the smallest enveloped animal viruses with a virion diameter of 42 nm, but pleomorphic forms exist, including filamentous and spherical bodies lacking a core. These particles are not infectious and are composed of the lipid and protein that forms part of the surface of the virion, which is called the surface antigen (HBsAg), and is produced in excess during the life cycle of the virus.[14]
Components
[edit]It consists of:
- HBsAg (hepatitis B surface antigen) was the first hepatitis B virus protein to be discovered.[15] It consists of small (S), medium (M) and large (L) protein.[16]
- HBcAg (hepatitis B core antigen) is the main structural protein of HBV icosahedral nucleocapsid and it has function in replication of the virus.[17] Capsid formation is the main factor for infection of the cell.[18] HBcAg contributes to HBV clearance in vivo, but it is unknown whether HBcAg has to be in the capsid form to contribute to viral clearance.[19]
- Hepatitis B virus DNA polymerase is incorporated into the nucleocapsid along with the pre-genomic RNA (pgRNA). Inside the capsid, the pgRNA undergoes reverse transcription, making the (-) DNA strand. At the same time, most of the RNA template is degraded by the RNase activity of the polymerase. This is followed by (+) DNA strand synthesis, and the polymerase ends up covalently bound to the (-) DNA strand.[20][21] The polymerase is discarded after the virion infects a new cell.
- HBeAg (hepatitis B envelope antigen) can be found between the icosahedral nucleocapsid core and the lipid envelope, but is considered "nonparticulate" and is secreted and accumulates in serum. HBeAg and HBcAg are made from the same reading frame.[22]
- HBx is small,[23] 154 amino acids long, nonstructural and has an important role in HBV-associated liver disease and in HBV replication in HepG2 cells. Many activities have been linked to expression of HBx. However, the molecular mechanisms of many of these activities are unknown.[24] This protein is multifunctional and it activates cellular signaling pathways and is essential for viral infection.[25]
Hepatitis D virus requires HBV envelope particles to become virulent.[26]
Evolution
[edit]The early evolution of HBV, like that of all viruses, is difficult to establish. The identification of hepadnaviruses in a wide range of vertebrates suggests a long coevolution. The identification of endogenous hepadnaviridae elements shared by various bird species shows the presence of these virus in birds for at least 70M years.[27] Although similar evidence is missing for mammals, the phylogenetic position of orthohepadnaviruses as a sister clade to avihepadnaviruses suggests a presence of the virus in the amniote ancestor and a subsequent coevolution with both birds and mammals after their divergence (>300M years ago). It has also been proposed that a New World bat hepadnavirus may be the origin of the primate hepadnaviruses.[28] Avihepadnaviruses lack the X protein but a vestigial X reading frame is present in the genome of duck hepadnavirus.[29] The X protein may have evolved from a DNA glycosylase.
Recently, the reconstruction of HBV genomes from ancient human remains has allowed investigating the evolution of this virus in humans in more details.[30][31][32] In 2021, a study reconstructed 137 ancient HBV genomes and proved the presence of the virus in humans since at least 10,000 years.[30] The most recent common ancestor of all known human HBV lineages was dated to between 20,000 and 12,000 years ago. However, it cannot be said whether the virus was present in humans long before that or acquired shortly before from another animal species. The evolution of HBV in humans was shown to reflect known events of human history such as the first peopling of the Americas during the late Pleistocene and the Neolithic transition in Europe.[30] These studies also showed that some ancient HBV strains still infect humans, while other became extinct.[30][31][32] HBV strains found in African and South-East Asian apes (chimpanzees, gorillas, orangutans, bonobos and gibbons) appear related to human HBV strains, which could reflect past cross-species transmission events.[33][30]
A study of isolates from the circumpolar Arctic human population has proposed that the ancestor of the subgenotype B5 (the endemic type found in this population) that the ancestral virus originated in Asia about 2000 years ago (95% HPD 900 BC – 830 AD).[34] Coalescence occurred about 1000 AD. This subgenotype spread from Asia initially to Greenland and then spread westward within the last 400 years.
Genome
[edit]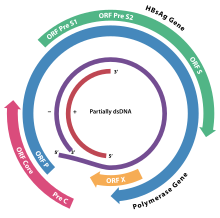
Size
[edit]The genome of HBV is made of circular DNA, but it is unusual because the DNA is not fully double-stranded. One end of the full length strand is linked to the viral DNA polymerase. The genome is 3020–3320 nucleotides long (for the full length strand) and 1700–2800 nucleotides long (for the short length strand).[35]
Encoding
[edit]The negative-sense, (non-coding) strand is complementary to the viral mRNA. The viral DNA is found in the nucleus soon after infection of the cell. The partially double-stranded DNA is rendered fully double-stranded by completion of the (+) sense strand by cellular DNA polymerases (viral DNA polymerase is used for a later stage) and removal of the viral polymerase protein (P) from the (−) sense strand and a short sequence of RNA from the (+) sense strand. Non-coding bases are removed from the ends of the (−)sense strand and the ends are rejoined.
The viral genes are transcribed by the cellular RNA polymerase II in the cell nucleus from a covalently closed circular DNA (cccDNA) template. Two enhancers designated enhancer I (EnhI) and enhancer II (EnhII) have been identified in the HBV genome. Both enhancers exhibit greater activity in cells of hepatic origin, and together they drive and regulate the expression of the complete viral transcripts.[36][37][38]
There are four known genes encoded by the genome called C, P, S, and X. The core protein is coded for by gene C (HBcAg), and its start codon is preceded by an upstream in-frame AUG start codon from which the pre-core protein is produced. HBeAg is produced by proteolytic processing of the pre-core protein. The DNA polymerase is encoded by gene P. Gene S is the gene that codes for the surface antigen (HBsAg). The HBsAg gene is one long open reading frame but contains three in frame "start" (ATG) codons that divide the gene into three sections, pre-S1, pre-S2, and S. Because of the multiple start codons, polypeptides of three different sizes called large (pre-S1 + pre-S2 + S), middle (pre-S2 + S), and small (S) are produced.[39]
The function of the protein coded for by gene X is not fully understood,[40] but some evidence suggests that it may function as a transcriptional transactivator. Interestingly, a 40 kDa X-Core fusion protein is encoded by a long viral 3.9-kb transcript, whose function remains unclear.[41] Synthesis of the 3.9 kb RNA initiates at the X gene promoter region and the transcript is polyadenylated only after the second round of transcription. Similar behavior is shared by other long pregenomic/pre-core (pg/pc) RNA species. Thus, the viral transcription machinery must ignore the poly(A) signal at the first transcription round.
Several non-coding RNA elements have been identified in the HBV genome. These include HBV PREalpha, HBV PREbeta and HBV RNA encapsidation signal epsilon.[42][43]
Genotypes
[edit]Genotypes differ by at least 8% of the sequence and have distinct geographical distributions and this has been associated with anthropological history. Within genotypes subtypes have been described: these differ by 4–8% of the genome.
There are eight known genotypes labeled A through H.[7]
A possible new "I" genotype has been described,[44] but acceptance of this notation is not universal.[45]
Two further genotypes have since been recognised.[46] The current (2014) listing now runs A through to J. Several subtypes are also recognised.
There are at least 24 subtypes.
Different genotypes may respond to treatment in different ways.[47][48]
- Individual genotypes
Type F which diverges from the other genomes by 14% is the most divergent type known. Type A is prevalent in Europe, Africa and South-east Asia, including the Philippines. Type B and C are predominant in Asia; type D is common in the Mediterranean area, the Middle East and India; type E is localized in sub-Saharan Africa; type F (or H) is restricted to Central and South America. Type G has been found in France and Germany. Genotypes A, D and F are predominant in Brazil and all genotypes occur in the United States with frequencies dependent on ethnicity.
The E and F strains appear to have originated in aboriginal populations of Africa and the New World, respectively.
Type A has two subtypes: Aa (A1) in Africa/Asia and the Philippines and Ae (A2) in Europe/United States.
Type B has two distinct geographical distributions: Bj/B1 ('j'—Japan) and Ba/B2 ('a'—Asia). Type Ba has been further subdivided into four clades (B2–B4).
Type C has two geographically subtypes: Cs (C1) in South-east Asia and Ce (C2) in East Asia. The C subtypes have been divided into five clades (C1–C5). A sixth clade (C6) has been described in the Philippines but only in one isolate to date.[49] Type C1 is associated with Vietnam, Myanmar and Thailand; type C2 with Japan, Korea and China; type C3 with New Caledonia and Polynesia; C4 with Australia; and C5 with the Philippines. A further subtype has been described in Papua, Indonesia.[50]
Type D has been divided into 7 subtypes (D1–D7).
Type F has been subdivided into 4 subtypes (F1–F4). F1 has been further divided into 1a and 1b. In Venezuela subtypes F1, F2, and F3 are found in East and West Amerindians. Among South Amerindians only F3 was found. Subtypes Ia, III, and IV exhibit a restricted geographic distribution (Central America, the North and the South of South America respectively) while clades Ib and II are found in all the Americas except in the Northern South America and North America respectively.
Life cycle
[edit]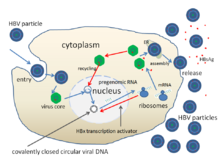
The life cycle of hepatitis B virus is complex. Hepatitis B is one of a few known non-retroviral viruses which use reverse transcription as a part of its replication process.
- Attachment
- The virus gains entry into the cell by binding to receptors on the surface of the cell and entering it by endocytosis mediated by either clathrin or caveolin-1.[51] HBV initially binds to heparin sulfate proteoglycan. The pre-S1 segment of the HBV L protein then binds tightly to the cell surface receptor sodium taurocholate cotransporting polypeptide (NTCP), encoded by the SLC10A1 gene.[52] NTCP is mostly found in the sinusoidal membrane of liver cells. The presence of NTCP in liver cells correlates with the tissue specificity of HBV infection.[51]
- Penetration
- Following endocytosis, the virus membrane fuses with the host cell's membrane, releasing the nucleocapsid into the cytoplasm.[53]
- Uncoating
- Because the virus multiplies via RNA made by a host enzyme, the viral genomic DNA has to be transferred to the cell nucleus. It is thought the capsid is transported on the microtubules to the nuclear pore. The core proteins dissociate from the partially double stranded viral DNA, which is then made fully double stranded (by host DNA polymerases) and transformed into covalently closed circular DNA (cccDNA) that serves as a template for transcription of four viral mRNAs.
- Replication
- The largest mRNA, (which is longer than the viral genome), is used to make the new copies of the genome and to make the capsid core protein and the viral RNA-dependent-DNA-polymerase.
- Assembly
- These four viral transcripts undergo additional processing and go on to form progeny virions which are released from the cell or returned to the nucleus and re-cycled to produce even more copies.[39][54]
- Release
- The long mRNA is then transported back to the cytoplasm where the virion P protein synthesizes DNA via its reverse transcriptase activity.
Disease
[edit]Despite there being a vaccine to prevent hepatitis B, HBV remains a global health problem. Hepatitis B can be acute and later become chronic, leading to other diseases and health conditions.[55] In addition to causing hepatitis, infection with HBV can lead to cirrhosis and hepatocellular carcinoma.[56]
It has also been suggested that it may increase the risk of pancreatic cancer.[4]
Roles in disease
[edit]Viral infection by hepatitis B virus (HBV) causes many hepatocyte changes due to the direct action of a protein encoded by the virus, HBx, and to indirect changes due to a large increase in intracellular reactive oxygen species (ROS) after infection. HBx appears to dysregulate a number of cellular pathways. HBx causes dysregulation in part by binding to genomic DNA, changing expression patterns of miRNAs, affecting histone methyltransferases, binding to SIRT1 protein to activate transcription, and cooperating with histone methylases and demethylases to change cell expression patterns.[57] HBx is partly responsible for the approximate 10,000-fold increase in intracellular reactive oxygen species (ROS) upon chronic HBV infection.[citation needed] Increased ROS can be caused, in part, by localization of HBx to the mitochondria where HBx decreases the mitochondrial membrane potential.[58] In addition, another HBV protein, HBsAg, also increases ROS through interactions with the endoplasmic reticulum.[58]
The increase in ROS after HBV infection causes inflammation, which leads to a further increase in ROS.[citation needed] ROS cause more than 20 types of DNA damage.[59] Oxidative DNA damage is mutagenic.[60] In addition, repair of the DNA damage can cause epigenetic alterations at the site of the damage during repair of the DNA.[61] Epigenetic alterations and mutations may cause defects in the cellular machinery that then contribute to liver disease. By the time accumulating epigenetic and mutational changes eventually cause progression to cancer, epigenetic alterations appear to have a larger role in this carcinogenesis than mutations. Only one or two genes, TP53[62] and perhaps ARID1A,[63] are mutated in more than 20% of liver cancers while 41 genes each have hypermethylated promoters (repressing gene expression) in more than 20% of liver cancers, with seven of these genes being hypermethylated in more than 75% of liver cancers.[62] In addition to alterations at the sites of DNA repair, epigenetic alterations are also caused by HBx recruiting the DNA methyltransferase enzymes, DNMT1 and/or DNMT3A, to specific gene loci to alter their methylation levels and gene expression.[64] HBx also alters histone acetylation that can affect gene expression.[64]
Several thousand protein-coding genes appear to have HBx-binding sites.[57][65] In addition to protein coding genes, about 15 microRNAs and 16 Long non-coding RNAs are also affected by the binding of HBx to their promoters.[65] Each altered microRNA can affect the expression of several hundred messenger RNAs (see microRNA).
History
[edit]The origin of hepatitis B virus can be traced back to the 5th century BCE and is even mentioned in Babylonian clay tablets. Hippocrates later described an epidemic of jaundice among his patients that was characterized by the yellowing of the skin and whites of the eyes. Jaundice is a clinical sign of hepatitis B viral infection.[66][67] However, due to the lengthy time interval, measured in weeks, between exposure of the causative agent and the development of illness prevented recognition of jaundice as an infectious disease until the 20th century.[68] The first recorded cases of hepatitis B virus infection occurred in 1883 after the smallpox vaccine containing human lymph was administered to a group of people.[68] The smallpox vaccine was administered to shipyard workers in Germany and the workers later developed symptoms of hepatitis.[68] Serum hepatitis, now known as hepatitis B, was often observed following the use of contaminated needles and syringes. These contaminated needles and syringes were not properly cleaned and/or they were reused among patients.[69] In 1943, transmission of hepatitis B virus via blood was further emphasized when Paul Beeson described jaundice occurring in patients who had just received blood transfusions. Another epidemic of jaundice was observed among soldiers in 1942, after they had received a yellow fever vaccine.[69] The distinction between hepatitis A virus and hepatitis B virus was not determined until 1947 when they were recognized as being two different filterable agents through numerous studies done of human volunteers.[69]
In 1964, the "Australian Antigen" was discovered by Barry Blumberg[70] and later identified as the hepatitis B virus surface antigen HBsAg. This was one of the first breakthroughs in the effort to understand the pathology of viral hepatitis that instigated jaundice in those infected with HBV. It allowed industrialized countries to reliably diagnose asymptomatic carriers of hepatitis B virus and the discovery provided healthcare professionals a way to screen blood for Hep B before administering blood transfusions.[69][68]
Today, hepatitis B virus infection is easily avoided by receiving one of the hepatitis B vaccines. The plasma-derived HepB vaccine was licensed in 1981 and was subsequently replaced in 1986 with the recombinant HepB vaccine. Engerix B was approved in 1989 and Heplisav-B was approved in 2017.[71][72][68] All of which provide protection against HBV.
Distribution
[edit]Rates of hepatitis B infection are equal across male and females. Hepatitis B virus is more prominently found in US citizens of Asian, Pacific Islander, or African descent and roughly 25% of these individuals will receive a diagnosis.[73] HBV is spread more readily in groups with high risk behavior such as Intravenous drug use, multiple sex partners, and men who have sex with men.[74]
Hepatitis B virus causes the disease hepatitis B. Hepatitis is considered to be the leading cause of liver cancer worldwide (reference). Hepatitis B virus can be found in almost every region of the world but is most prevalent in countries where the virus is endemic. HBV is endemic in some countries located in Asia, Africa, South America, and the Caribbean.[75]
Approximately two billion people have been infected with HBV which means almost 1 out of 3 people have been infected. Every year an estimated 1.5 million people will become newly infected and roughly 10% of those individuals will go undiagnosed. Every year, an estimated 820,000 people die from hepatitis B infection and related HBV complications.[76]
The spread of HBV during pregnancy remains the highest risk for developing chronic hepatitis B later in childhood. Roughly 90% of infected infants will become chronically infected. Only 2%-6% of adults once infected with HBV will go on to be chronically infected.[77] Of the estimated 350 million individuals chronically infected with HBV worldwide, 50% or more of those individuals acquired the infection prenatally or during their early childhood. In countries where HBV is endemic, vertical transmission of HBV poses a major health risk due to a high number of women of childbearing age being HBeAg-positive allowing them to transmit HBV to their newborn. In areas where HBV is endemic, transmission is not limited to groups with high-risk behaviors. Instead, infection can occur through different routes of transmission but mostly during early childhood.[78]
The spread of hepatitis B virus in the western world occurs most often through sexual intercourse or needle sharing by intravenous drug users (IVDU). IVDU show the highest rate of HBV infection in Europe and North America.[74] There are also higher rates of hepatitis B infection among men who have sex with men (MSM). Risk of being infected with HBV increases with having multiple sex partners. (need reference)
Transmission
[edit]The spread of hepatitis B virus occurs most often through vertical transmission from mother to child during birth and delivery. HBV can also be spread through contact with blood or other bodily fluids during sexual intercourse with an infected partner. It is also spread through needles shared with infected persons or exposure to sharp objects. Needles of any kind can be a risk if they are not single use or are not properly sanitized, preferably in an autoclave. This includes needles used at tattooing and body piercing parlors.[75]
Furthermore, hepatitis B virus can also be spread through sharing earrings and other body piercing jewelry.[76] It is also spread through hemodialysis units that have been used by HBeAg positive patients. Because HD units usually treat multiple patients at a time, contamination of patients' blood can occur. Incidences of HBV infection through HD units is at 1% in the United States. Healthcare staff are also at an increased risk of infection.[79][80] Transmission of HBV can be limited by administering the hepatitis B vaccine. In areas where the virus is endemic, vaccines are limited, especially in rural areas where medical clinics are sparse.
Although HBV can be infectious on surfaces for up to seven days, it is not spread through breastfeeding, sharing eating utensils, hugging, kissing, holding hands, coughing, or sneezing. Unlike other hepatitis viruses, HBV is not spread by contaminated food or water. However, living with a person infected with hepatitis B virus increases your risk of contracting the virus.[68]
Co-infection of HBV and other viruses
[edit]Co-infection of hepatitis B and various other viruses can also occur. hepatitis C, hepatitis D (a satellite virus of hepatitis B), and HIV can all co-infect an individual alongside HBV.
Because HBV and HCV share a similar mode of transmission, co-infections are possible. Most cases of HBV and HCV co-infection occurs among Intravenous drug users, unscreened blood products, or exposure to dirty needles and unsterilized medical equipment. Co-infection of these two viruses can cause a more severe liver disease and increase the risk for primary liver cancer (Hepatocellular Carcinoma). Reporting of this co-infection may be underreported due to hepatitis C's ability to become the dominant liver virus during coinfection, reducing the detectable amount of HBV found in the body.[81] Recent statistics show that 10% of all persons infected with HIV are also infected with Hepatitis B. However, this statistic increases to almost 20% for Southeast Asia. Hepatitis B infection is one of the leading causes of hospitalizations and death among patients with HIV since the development and use of antiretroviral therapies. Those who are infected with HIV and HBV are six times more likely to develop chronic hepatitis B. Some studies suggest this may be due to co-infected individuals having lower CD4+ T cell counts.[82]
See also
[edit]- Hepatitis B vaccine
- Nucleoside analogues
- Oncovirus (cancer virus)
References
[edit]- ^ Ryu WS (2017). Molecular Virology of Human Pathogenic Viruses. Academic Press. pp. 247–260. ISBN 978-0-12-800838-6.
- ^ Hunt R (21 November 2007). "Hepatitis viruses". University of Southern California, Department of Pathology and Microbiology. Retrieved 13 March 2008.
- ^ a b c d "ICTV Report Hepadnaviridae".
- ^ a b Hassan MM, Li D, El-Deeb AS, Wolff RA, Bondy ML, Davila M, Abbruzzese JL (October 2008). "Association between hepatitis B virus and pancreatic cancer". Journal of Clinical Oncology. 26 (28): 4557–62. doi:10.1200/JCO.2008.17.3526. PMC 2562875. PMID 18824707.
- ^ Dupinay T, et al. (November 2013). "Discovery of naturally occurring transmissible chronic hepatitis B virus infection among Macaca fascicularis from Mauritius Island". Hepatology. 58 (5): 1610–1620. doi:10.1002/hep.26428. PMID 23536484. S2CID 205888844.
- ^ a b Hundie GB, Stalin Raj V, Gebre Michael D, Pas SD, Koopmans MP, Osterhaus AD, et al. (February 2017). "A novel hepatitis B virus subgenotype D10 circulating in Ethiopia". Journal of Viral Hepatitis. 24 (2): 163–173. doi:10.1111/jvh.12631. PMID 27808472. S2CID 23073883.
- ^ a b Kramvis A, Kew M, François G (March 2005). "Hepatitis B virus genotypes". Vaccine. 23 (19): 2409–23. doi:10.1016/j.vaccine.2004.10.045. PMID 15752827.
- ^ Magnius LO, Norder H (1995). "Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene". Intervirology. 38 (1–2): 24–34. doi:10.1159/000150411. PMID 8666521.
- ^ Ghosh S, Banerjee P, Deny P, Mondal RK, Nandi M, Roychoudhury A, et al. (March 2013). "New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India". Journal of Viral Hepatitis. 20 (3): 209–18. doi:10.1111/j.1365-2893.2012.01655.x. PMID 23383660. S2CID 205356299.
- ^ Drexler JF, Geipel A, König A, Corman VM, van Riel D, Leijten LM, et al. (October 2013). "Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes". Proceedings of the National Academy of Sciences of the United States of America. 110 (40): 16151–6. Bibcode:2013PNAS..11016151D. doi:10.1073/pnas.1308049110. PMC 3791787. PMID 24043818.
- ^ Zuckerman AJ (1996). "Chapter 70: Hepatitis Viruses". In Baron S; et al. (eds.). Baron's Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. Retrieved 11 April 2018.
- ^ "WHO | Hepatitis B". www.who.int. Archived from the original on 10 July 2015. Retrieved 12 July 2015.
- ^ Locarnini S (2004). "Molecular virology of hepatitis B virus". Seminars in Liver Disease. 24 (Suppl 1): 3–10. CiteSeerX 10.1.1.618.7033. doi:10.1055/s-2004-828672. PMID 15192795. S2CID 260320531.
- ^ Howard CR (July 1986). "The biology of hepadnaviruses". The Journal of General Virology. 67 (7): 1215–35. doi:10.1099/0022-1317-67-7-1215. PMID 3014045.
- ^ Jaroszewicz J, Calle Serrano B, Wursthorn K, Deterding K, Schlue J, Raupach R, et al. (April 2010). "Hepatitis B surface antigen (HBsAg) levels in the natural history of hepatitis B virus (HBV)-infection: a European perspective". Journal of Hepatology. 52 (4): 514–22. doi:10.1016/j.jhep.2010.01.014. PMID 20207438.
- ^ Seeger C, Mason WS (March 2000). "Hepatitis B virus biology". Microbiology and Molecular Biology Reviews. 64 (1): 51–68. doi:10.1128/mmbr.64.1.51-68.2000. PMC 98986. PMID 10704474.
- ^ Lin YJ, Wu HL, Chen DS, Chen PJ (September 2012). "Hepatitis B virus nucleocapsid but not free core antigen controls viral clearance in mice". Journal of Virology. 86 (17): 9266–73. doi:10.1128/JVI.00608-12. PMC 3416136. PMID 22718814.
- ^ Lin YJ, Huang LR, Yang HC, Tzeng HT, Hsu PN, Wu HL, et al. (May 2010). "Hepatitis B virus core antigen determines viral persistence in a C57BL/6 mouse model". Proceedings of the National Academy of Sciences of the United States of America. 107 (20): 9340–5. Bibcode:2010PNAS..107.9340L. doi:10.1073/pnas.1004762107. PMC 2889105. PMID 20439715.
- ^ Bourne CR, Katen SP, Fulz MR, Packianathan C, Zlotnick A (March 2009). "A mutant hepatitis B virus core protein mimics inhibitors of icosahedral capsid self-assembly". Biochemistry. 48 (8): 1736–42. doi:10.1021/bi801814y. PMC 2880625. PMID 19196007.
- ^ Menéndez-Arias L, Álvarez M, Pacheco B (October 2014). "Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: mechanism of action and resistance". Current Opinion in Virology. 8: 1–9. doi:10.1016/j.coviro.2014.04.005. PMID 24814823.
- ^ Yang HC, Kao JH (September 2014). "Persistence of hepatitis B virus covalently closed circular DNA in hepatocytes: molecular mechanisms and clinical significance". Emerging Microbes & Infections. 3 (9): e64. doi:10.1038/emi.2014.64. PMC 4185362. PMID 26038757.
- ^ "TSRI - News and Publications". Retrieved 3 January 2009.
- ^ Tang H, Oishi N, Kaneko S, Murakami S (October 2006). "Molecular functions and biological roles of hepatitis B virus x protein". Cancer Science. 97 (10): 977–83. doi:10.1111/j.1349-7006.2006.00299.x. PMC 11159107. PMID 16984372.
- ^ McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ (November 2007). "Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels". Journal of Virology. 81 (21): 12061–5. doi:10.1128/JVI.00740-07. PMC 2168786. PMID 17699583.
- ^ Bouchard MJ, Puro RJ, Wang L, Schneider RJ (July 2003). "Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication". Journal of Virology. 77 (14): 7713–9. doi:10.1128/JVI.77.14.7713-7719.2003. PMC 161925. PMID 12829810.
- ^ Chai N, Chang HE, Nicolas E, Han Z, Jarnik M, Taylor J (August 2008). "Properties of subviral particles of hepatitis B virus". Journal of Virology. 82 (16): 7812–7. doi:10.1128/JVI.00561-08. PMC 2519590. PMID 18524834.
- ^ Suh, Alexander; Brosius, Jürgen; Schmitz, Jürgen; Kriegs, Jan Ole (30 April 2013). "The genome of a Mesozoic paleovirus reveals the evolution of hepatitis B viruses". Nature Communications. 4 (1): 1791. Bibcode:2013NatCo...4.1791S. doi:10.1038/ncomms2798. ISSN 2041-1723. PMID 23653203.
- ^ Rasche A, Souza BF, Drexler JF (February 2016). "Bat hepadnaviruses and the origins of primate hepatitis B viruses". Current Opinion in Virology. 16: 86–94. doi:10.1016/j.coviro.2016.01.015. PMID 26897577.
- ^ Lin B, Anderson DA (2000). "A vestigial X open reading frame in duck hepatitis B virus". Intervirology. 43 (3): 185–90. doi:10.1159/000025037. PMID 11044813. S2CID 22542029.
- ^ a b c d e Kocher, Arthur; Papac, Luka; Barquera, Rodrigo; Key, Felix M.; Spyrou, Maria A.; Hübler, Ron; Rohrlach, Adam B.; Aron, Franziska; Stahl, Raphaela; Wissgott, Antje; Bömmel, Florian van (8 October 2021). "Ten millennia of hepatitis B virus evolution". Science. 374 (6564): 182–188. Bibcode:2021Sci...374..182K. doi:10.1126/science.abi5658. hdl:1887/3256616. PMID 34618559. S2CID 238475573.
- ^ a b Mühlemann B, Jones TC, Damgaard PB, Allentoft ME, Shevnina I, Logvin A, et al. (May 2018). "Ancient hepatitis B viruses from the Bronze Age to the Medieval period". Nature. 557 (7705): 418–423. Bibcode:2018Natur.557..418M. doi:10.1038/s41586-018-0097-z. PMID 29743673. S2CID 13684815.
- ^ a b Krause-Kyora, Ben; Susat, Julian; Key, Felix M; Kühnert, Denise; Bosse, Esther; Immel, Alexander; Rinne, Christoph; Kornell, Sabin-Christin; Yepes, Diego; Franzenburg, Sören; Heyne, Henrike O (10 May 2018). Locarnini, Stephen (ed.). "Neolithic and medieval virus genomes reveal complex evolution of hepatitis B". eLife. 7: e36666. doi:10.7554/eLife.36666. ISSN 2050-084X. PMC 6008052. PMID 29745896.
- ^ Paraskevis D, Magiorkinis G, Magiorkinis E, Ho SY, Belshaw R, Allain JP, Hatzakis A (March 2013). "Dating the origin and dispersal of hepatitis B virus infection in humans and primates". Hepatology. 57 (3): 908–16. doi:10.1002/hep.26079. PMID 22987324.
- ^ Bouckaert R, Simons BC, Krarup H, Friesen TM, Osiowy C (2017). "Tracing hepatitis B virus (HBV) genotype B5 (formerly B6) evolutionary history in the circumpolar Arctic through phylogeographic modelling". PeerJ. 5: e3757. doi:10.7717/peerj.3757. PMC 5581946. PMID 28875087.
- ^ Kay A, Zoulim F (August 2007). "Hepatitis B virus genetic variability and evolution". Virus Research. 127 (2): 164–76. doi:10.1016/j.virusres.2007.02.021. PMID 17383765.
- ^ Doitsh G, Shaul Y (February 2004). "Enhancer I predominance in hepatitis B virus gene expression". Molecular and Cellular Biology. 24 (4): 1799–808. doi:10.1128/mcb.24.4.1799-1808.2004. PMC 344184. PMID 14749394.
- ^ Antonucci TK, Rutter WJ (February 1989). "Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner". Journal of Virology. 63 (2): 579–83. doi:10.1128/JVI.63.2.579-583.1989. PMC 247726. PMID 2536093.
- ^ Huan B, Siddiqui A (1993). "Regulation of hepatitis B virus gene expression". Journal of Hepatology. 17 (Suppl 3): S20-3. doi:10.1016/s0168-8278(05)80419-2. PMID 8509635.
- ^ a b Beck J, Nassal M (January 2007). "Hepatitis B virus replication". World Journal of Gastroenterology. 13 (1): 48–64. doi:10.3748/wjg.v13.i1.48. PMC 4065876. PMID 17206754.
- ^ Bouchard MJ, Schneider RJ (December 2004). "The enigmatic X gene of hepatitis B virus". Journal of Virology. 78 (23): 12725–34. doi:10.1128/JVI.78.23.12725-12734.2004. PMC 524990. PMID 15542625.
- ^ Doitsh, Gilad; Shaul, Yosef (May 2003). "A long HBV transcript encoding pX is inefficiently exported from the nucleus". Virology. 309 (2): 339–349. doi:10.1016/S0042-6822(03)00156-9. PMID 12758180.
- ^ Smith GJ, Donello JE, Lück R, Steger G, Hope TJ (November 1998). "The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function". Nucleic Acids Research. 26 (21): 4818–27. doi:10.1093/nar/26.21.4818. PMC 147918. PMID 9776740.
- ^ Flodell S, Schleucher J, Cromsigt J, Ippel H, Kidd-Ljunggren K, Wijmenga S (November 2002). "The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges". Nucleic Acids Research. 30 (21): 4803–11. doi:10.1093/nar/gkf603. PMC 135823. PMID 12409471.
- ^ Olinger CM, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, et al. (November 2008). "Possible new hepatitis B virus genotype, southeast Asia". Emerging Infectious Diseases. 14 (11): 1777–80. doi:10.3201/eid1411.080437. PMC 2630741. PMID 18976569.
- ^ Kurbanov F, Tanaka Y, Kramvis A, Simmonds P, Mizokami M (August 2008). "When should "I" consider a new hepatitis B virus genotype?". Journal of Virology. 82 (16): 8241–2. doi:10.1128/JVI.00793-08. PMC 2519592. PMID 18663008.
- ^ Hernández S, Venegas M, Brahm J, Villanueva RA (October 2014). "Full-genome sequence of a hepatitis B virus genotype f1b clone from a chronically infected chilean patient". Genome Announcements. 2 (5): e01075–14. doi:10.1128/genomeA.01075-14. PMC 4208329. PMID 25342685.
- ^ Palumbo E (2007). "Hepatitis B genotypes and response to antiviral therapy: a review". American Journal of Therapeutics. 14 (3): 306–9. doi:10.1097/01.pap.0000249927.67907.eb. PMID 17515708. S2CID 35134058.
- ^ Mahtab MA, Rahman S, Khan M, Karim F (October 2008). "Hepatitis B virus genotypes: an overview". Hepatobiliary & Pancreatic Diseases International. 7 (5): 457–64. PMID 18842489.
- ^ Cavinta L, Sun J, May A, Yin J, von Meltzer M, Radtke M, et al. (June 2009). "A new isolate of hepatitis B virus from the Philippines possibly representing a new subgenotype C6". Journal of Medical Virology. 81 (6): 983–7. doi:10.1002/jmv.21475. PMID 19382274. S2CID 23251719.
- ^ Lusida MI, Nugrahaputra VE, Handajani R, Nagano-Fujii M, Sasayama M, Utsumi T, Hotta H (July 2008). "Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia". Journal of Clinical Microbiology. 46 (7): 2160–6. doi:10.1128/JCM.01681-07. PMC 2446895. PMID 18463220.
- ^ a b Zhang Z, Zehnder B, Damrau C, Urban S (July 2016). "Visualization of hepatitis B virus entry - novel tools and approaches to directly follow virus entry into hepatocytes". FEBS Letters. 590 (13): 1915–26. doi:10.1002/1873-3468.12202. PMID 27149321.
- ^ Yan H, Liu Y, Sui J, Li W (September 2015). "NTCP opens the door for hepatitis B virus infection". Antiviral Research. 121: 24–30. doi:10.1016/j.antiviral.2015.06.002. PMID 26071008.
- ^ Watashi K, Wakita T (August 2015). "Hepatitis B Virus and Hepatitis D Virus Entry, Species Specificity, and Tissue Tropism". Cold Spring Harbor Perspectives in Medicine. 5 (8): a021378. doi:10.1101/cshperspect.a021378. PMC 4526719. PMID 26238794.
- ^ Bruss V (January 2007). "Hepatitis B virus morphogenesis". World Journal of Gastroenterology. 13 (1): 65–73. doi:10.3748/wjg.v13.i1.65. PMC 4065877. PMID 17206755.
- ^ Hu, J.; Protzer, U.; Siddiqui, A. (2019). "Revisiting Hepatitis B Virus: Challenges of Curative Therapies". Journal of Virology. 93 (20). doi:10.1128/JVI.01032-19. PMC 6798116. PMID 31375584.
- ^ Schwalbe M, Ohlenschläger O, Marchanka A, Ramachandran R, Häfner S, Heise T, Görlach M (March 2008). "Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element". Nucleic Acids Research. 36 (5): 1681–9. doi:10.1093/nar/gkn006. PMC 2275152. PMID 18263618.
- ^ a b Balakrishnan L, Milavetz B (November 2017). "Epigenetic Regulation of Viral Biological Processes". Viruses. 9 (11): 346. doi:10.3390/v9110346. PMC 5707553. PMID 29149060.
- ^ a b Higgs MR, Chouteau P, Lerat H (May 2014). "'Liver let die': oxidative DNA damage and hepatotropic viruses" (PDF). The Journal of General Virology. 95 (Pt 5): 991–1004. doi:10.1099/vir.0.059485-0. PMID 24496828.
- ^ Yu Y, Cui Y, Niedernhofer LJ, Wang Y (December 2016). "Occurrence, Biological Consequences, and Human Health Relevance of Oxidative Stress-Induced DNA Damage". Chemical Research in Toxicology. 29 (12): 2008–2039. doi:10.1021/acs.chemrestox.6b00265. PMC 5614522. PMID 27989142.
- ^ Dizdaroglu M (December 2012). "Oxidatively induced DNA damage: mechanisms, repair and disease". Cancer Letters. 327 (1–2): 26–47. doi:10.1016/j.canlet.2012.01.016. PMID 22293091.
- ^ Nishida N, Kudo M (2013). "Oxidative stress and epigenetic instability in human hepatocarcinogenesis". Digestive Diseases. 31 (5–6): 447–53. doi:10.1159/000355243. PMID 24281019.
- ^ a b Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U, et al. (May 2013). "Genetics and epigenetics of liver cancer". New Biotechnology. 30 (4): 381–4. doi:10.1016/j.nbt.2013.01.007. hdl:11693/20956. PMID 23392071.
- ^ Shibata T, Aburatani H (June 2014). "Exploration of liver cancer genomes". Nature Reviews. Gastroenterology & Hepatology. 11 (6): 340–9. doi:10.1038/nrgastro.2014.6. PMID 24473361. S2CID 8611393.
- ^ a b Tian Y, Yang W, Song J, Wu Y, Ni B (August 2013). "Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis". Molecular and Cellular Biology. 33 (15): 2810–6. doi:10.1128/MCB.00205-13. PMC 3719687. PMID 23716588.
- ^ a b Guerrieri F, Belloni L, D'Andrea D, Pediconi N, Le Pera L, Testoni B, et al. (February 2017). "Genome-wide identification of direct HBx genomic targets". BMC Genomics. 18 (1): 184. doi:10.1186/s12864-017-3561-5. PMC 5316204. PMID 28212627.
- ^ Liang, T. Jake (May 2009). "Hepatitis B: The virus and disease". Hepatology. 49 (S5): S13–S21. doi:10.1002/hep.22881. PMC 2809016. PMID 19399811.
- ^ "VA.gov | Veterans Affairs". www.hepatitis.va.gov. Retrieved 4 December 2022.
- ^ a b c d e f "Pinkbook: Hepatitis B | CDC". www.cdc.gov. 22 September 2022. Retrieved 3 December 2022.
- ^ a b c d Block, Timothy M.; Alter, Harvey J.; London, W. Thomas; Bray, Mike (1 July 2016). "A historical perspective on the discovery and elucidation of the hepatitis B virus". Antiviral Research. 131: 109–123. doi:10.1016/j.antiviral.2016.04.012. ISSN 0166-3542. PMID 27107897.
- ^ Blumberg, B. S. (1964). "Polymorphisms of the serum proteins and the development of iso-preciptins in transfused patients". Bull N Y Acad Med. 40 (5): 377–386. PMC 1750599. PMID 14146804.
- ^ Research, Center for Biologics Evaluation and (3 October 2019). "ENGERIX-B". FDA.
- ^ "Immunization Schedules for Heplisav-B Vaccine | CDC". www.cdc.gov. 8 June 2022. Retrieved 4 December 2022.
- ^ "Hepatitis B Facts and Figures". www.hepb.org. Retrieved 2 December 2022.
- ^ a b Piot, P.; Goilav, C.; Kegels, E. (1 March 1990). "Hepatitis B: transmission by sexual contact and needle sharing". Vaccine. 8: S37–S40. doi:10.1016/0264-410X(90)90215-8. ISSN 0264-410X. PMID 2183516.
- ^ a b "Hepatitis B | Disease Directory | Travelers' Health | CDC". wwwnc.cdc.gov. Retrieved 2 December 2022.
- ^ a b "Hepatitis B". www.who.int. Retrieved 2 December 2022.
- ^ "Safety Information for Hepatitis B Vaccines | Vaccine Safety | CDC". www.cdc.gov. 23 September 2021. Retrieved 4 December 2022.
- ^ Edmunds, W. J.; Medley, G. F.; Nokes, D. J.; O'Callaghan, C. J.; Whittle, H. C.; Hall, A. J. (1996). "Epidemiological Patterns of Hepatitis B Virus (HBV) in Highly Endemic Areas". Epidemiology and Infection. 117 (2): 313–325. doi:10.1017/S0950268800001497. ISSN 0950-2688. JSTOR 3864222. PMC 2271713. PMID 8870629.
- ^ Garthwaite, Elizabeth; Reddy, Veena; Douthwaite, Sam; Lines, Simon; Tyerman, Kay; Eccles, James (28 October 2019). "Clinical practice guideline management of blood borne viruses within the haemodialysis unit". BMC Nephrology. 20 (1): 388. doi:10.1186/s12882-019-1529-1. ISSN 1471-2369. PMC 6816193. PMID 31656166.
- ^ Fabrizi, Fabrizio; Dixit, Vivek; Messa, Piergiorgio; Martin, Paul (January 2015). "Transmission of Hepatitis B Virus in Dialysis Units: A Systematic Review of Reports on Outbreaks". The International Journal of Artificial Organs. 38 (1): 1–7. doi:10.5301/ijao.5000376. ISSN 0391-3988. PMID 25633894. S2CID 31944731.
- ^ "Hepatitis B Foundation: Hepatitis C and Hepatitis B Coinfection". www.hepb.org. Retrieved 2 December 2022.
- ^ Thio, Chloe L. (May 2009). "Hepatitis B and human immunodeficiency virus coinfection". Hepatology. 49 (S5): S138–S145. doi:10.1002/hep.22883. PMID 19399813. S2CID 26373510.
